Creates a forest plot from a `gtsummary`-style object produced by `gtregression` functions (e.g., `uni_reg()`, `multi_reg()`). The function supports both univariate and multivariable models, renders hierarchical labels (variable headers vs. levels), and computes significance highlighting using either *p*-values (linear models) or CI-vs-reference rules (non-linear models).
Usage
plot_reg(
tbl,
title = NULL,
order_y = NULL,
log_x = FALSE,
xlim = NULL,
breaks = NULL,
point_color = "#1F77B4",
errorbar_color = "#4C4C4C",
base_size = 14,
show_ref = TRUE,
sig_color = NULL,
sig_errorbar_color = NULL,
alpha = 0.05
)Arguments
- tbl
A `gtsummary`-like object returned by `gtregression` (must contain `table_body` and attributes `source` and `approach`).
- title
Optional plot title (character).
- order_y
Optional character vector to customize the y-axis header ordering.
- log_x
Logical. If `TRUE`, log x-axis (ignored for linear models).
- xlim
Optional numeric vector of length 2 for x-axis limits.
- breaks
Optional numeric vector for x-axis tick breaks (ignored if `log_x = TRUE`).
- point_color
Fill color for points (default `"#1F77B4"`).
- errorbar_color
Color for all error bars (default `"#4C4C4C"`).
- base_size
Base font size for `theme_minimal()` (default `14`).
- show_ref
Logical. If `TRUE`, includes the reference level on the plot and labels it `(Ref.)`.
- sig_color
Optional fill color for **significant** points; if `NULL`, significant points reuse `point_color`.
- sig_errorbar_color
Optional color for **significant** error bars; if `NULL`, significant bars reuse `errorbar_color`.
- alpha
Significance level for linear models when `p.value` is available (default `0.05`).
Details
**Reference line**: The vertical reference is fixed at `0` for linear models and `1` for all other approaches, inferred from `attr(tbl, "approach")`.
**Header / data detection**: Variable headers are recognized via `row_type == "label"` together with `header_row` or missing CI; categorical levels use `row_type == "level"`; continuous predictors appear as `row_type == "label"` **with** CIs and are treated as data rows.
**Significance highlighting**: - For `approach == "linear"` with available `p.value`, rows are significant when `p.value < alpha`. - Otherwise, rows are significant when the CI does not cross the reference (`0` or `1` as above). Use `sig_color` / `sig_errorbar_color` to customize the appearance.
Examples
# \donttest{
if (requireNamespace("mlbench", quietly = TRUE) &&
requireNamespace("gtregression", quietly = TRUE)) {
data("PimaIndiansDiabetes2", package = "mlbench")
pima <- PimaIndiansDiabetes2
pima$diabetes <- ifelse(pima$diabetes == "pos", 1, 0)
pima$bmi_cat <- cut(
pima$mass,
breaks = c(-Inf, 18.5, 24.9, 29.9, Inf),
labels = c("Underweight", "Normal", "Overweight", "Obese")
)
# Univariate logistic regression table via gtregression
tbl_uni <- gtregression::uni_reg(
data = pima,
outcome = "diabetes",
exposures = c("age", "bmi_cat"),
approach = "logit"
)
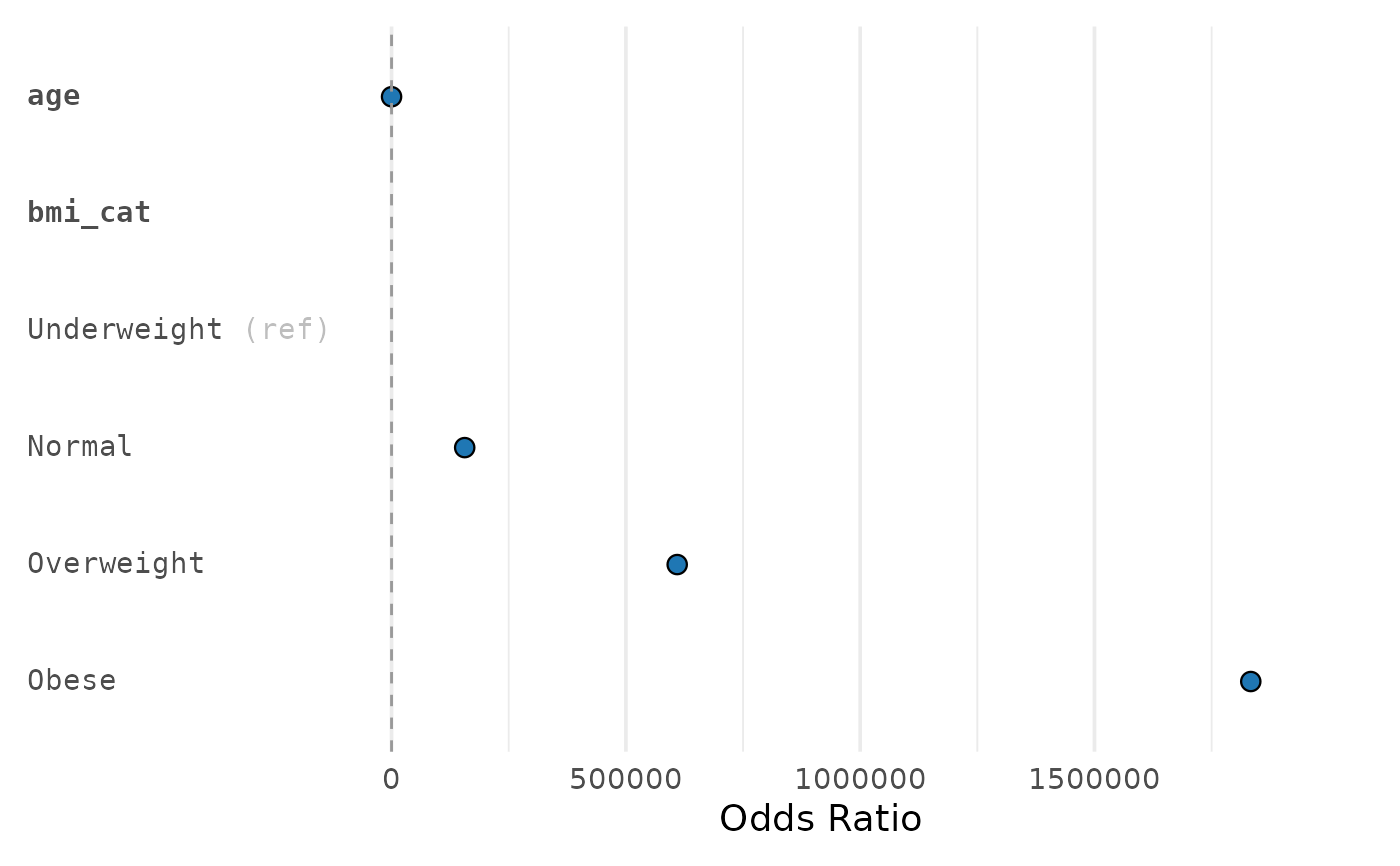
p <- plot_reg(tbl_uni, title = "Univariate (logit)", sig_color = "#D55E00")
print(p)
}
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: collapsing to unique 'x' values
 # }
# }